%%capture
! pip install -U ovitoRecipe 3: Color coding grain boundary atoms
How to Script with OVITO
When you have grain boundaries it is convenient to be able to distinguish those atoms from the bulk. One way to do so is using a order or structural parameter that captures some information about the local environment. OVITO has a few different options built in that enable characterize whether an atom is in an FCC, BCC, HCP, or other structure. Here I’ll be working with the LAMMPS dump file from the example in (Bringuier 2015) which reproduces the results from (Cahn, Mishin, and Suzuki 2006).
Import OVITO modules
from ovito.io import import_file
from ovito.vis import Viewport
from ovito.modifiers import CommonNeighborAnalysisModifier, CalculateDisplacementsModifier
from ovito.modifiers import ExpressionSelectionModifier, DeleteSelectedModifier, InvertSelectionModifier
from ovito.modifiers import AssignColorModifier
from ovito.vis import VectorVis
from ovito.vis import OSPRayRendererStep 1: Download and import file
This LAMMPS dump file was from a 2D simulation but the symmetry is 3D so we reset the PBC for all directions. I’m doing this because I want to maintain the correct structural analysis results.
%%capture
! wget 'https://drive.google.com/uc?id=1Id0D6rfHxJOTuPx2RVAYkm2yRBBgIzSE&export=download' -O dump.Cu_Bicrystal_Shear_298K.gz
pipeline = import_file('dump.Cu_Bicrystal_Shear_298K.gz',input_format='lammps/dump')
pipeline.source.data.cell_.pbc = (True, True, True)Step 2: Perform structural analysis and calculate displacement vectors
I will use the CommonNeighborAnalysisModifier which provides standard structural analysis to identify the closed-packing of the crystal (e.g., FCC, BCC, HCP, ICO).
Once the structure modifier is used, I will color code just the atoms that are of StructureType == which corresponds to “Other”. To do this we can use the modifier to select these atoms and then color code.
pipeline.modifiers.append(CommonNeighborAnalysisModifier())
# Select particles based on structure type and position
expression = 'StructureType != 0 || (Position.Z > CellSize.Z-5 || Position.Z < 5.0 )'
pipeline.modifiers.append(ExpressionSelectionModifier(expression=expression))
# Color the selected particles
pipeline.modifiers.append(AssignColorModifier(color=(0.0, 0.5, 1.0)))
pipeline.modifiers.append(InvertSelectionModifier())
# Now color the GB atoms
pipeline.modifiers.append(AssignColorModifier(color=(0.8, 0.5, 0.2)))
pipeline.modifiers.append(InvertSelectionModifier())
pipeline.add_to_scene()Step 3: Render first animation
vp = Viewport(type=Viewport.Type.Ortho)
vp.camera_dir = [-1, 0, 0]
vp.zoom_all()
renderer = OSPRayRenderer(
ambient_light_enabled=True,
denoising_enabled=True,
direct_light_enabled=True,
direct_light_intensity=1.0,
material_shininess=10.0,
material_specular_brightness=0.02
)
fname = "Cu_Bicrystal_Shear_298K_ColorCoding.gif"
vp.render_anim(size=(600,400),
filename=fname,
renderer=renderer)The code below is to allow for displaying rendered images in Google Colab.
try:
import google.colab
from IPython.display import Image
Image(open(fname, 'rb').read())
except ImportError:
"Assuming local run."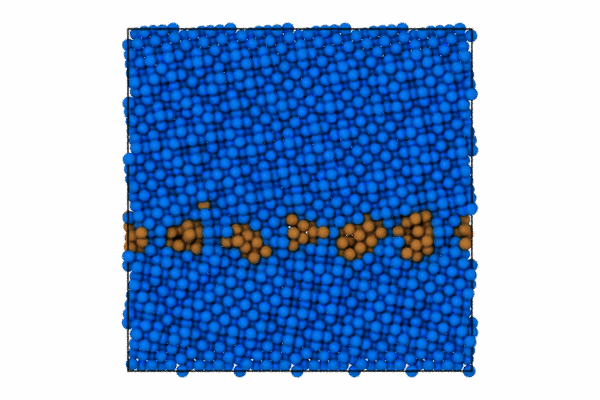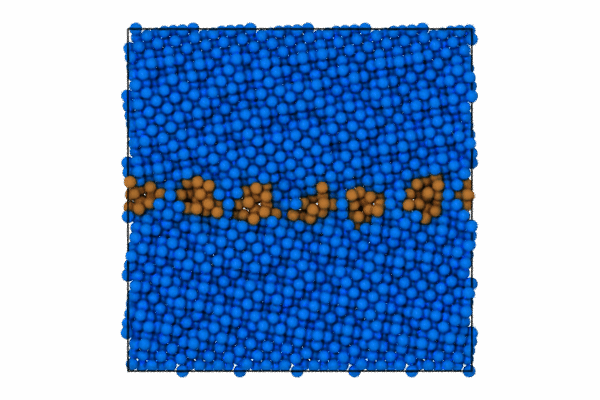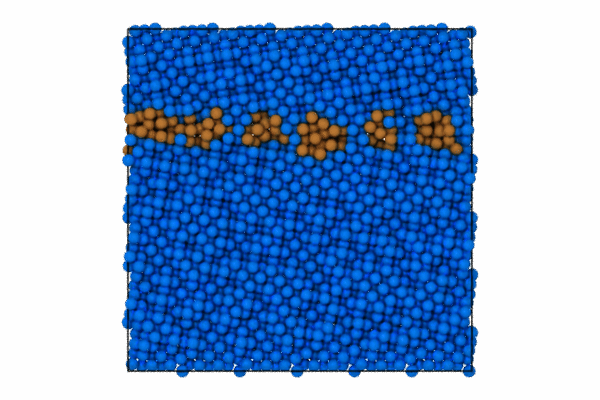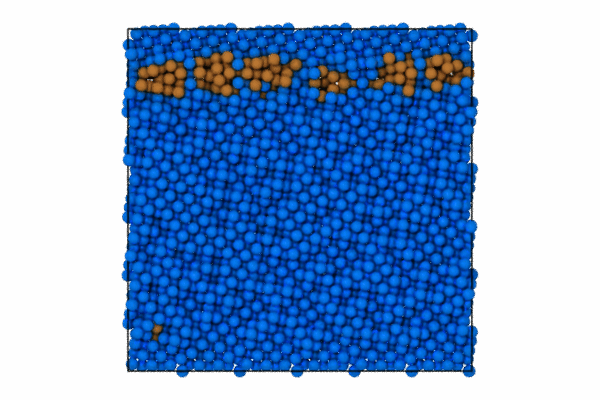
Step 4: Displacement vectors
We may want to show how the atoms at the grain boundary are moving with respect to the shear force. We can do this with the displacement vectors. The displacement vectors are taken with respect to an previous frame offset of 25. This is used to showcase how the grain boundary atoms are moving, what you’ll see is that its in a cork-screw like manner.
Since we already have a pipeline and all we want to do is add the displacement vectors and then disable visualizing the particles themselves. I also just want to so the displacement vectors for the grain boundary atoms, so I need to delete the selected particles and then we just need to add a few additional lines of code and then re-render to a new animation file.
# Visualization object for vector arrows
vec_vis = VectorVis(width=0.1,
scaling=1.5,
flat_shading=False,
color=(1,0,0))
displacement_mod = CalculateDisplacementsModifier(frame_offset=-25,
use_frame_offset=True,
vis=vec_vis)
displacement_mod.vis.enabled = True
pipeline.modifiers.append(displacement_mod)
# Need this line to turn off particle visibility
# Delete elected particles
pipeline.modifiers.append(DeleteSelectedModifier(operate_on={'particles'}))
pipeline.compute(frame=25).particles.vis.enabled = FalseStep 5: Render second animation
vp2 = Viewport(type=Viewport.Type.Ortho)
vp2.camera_dir = [-1, 0, 0]
vp2.zoom_all()
fname = "Cu_Bicrystal_Shear_298K_DisplacementVectors.gif"
nframes = pipeline.source.num_frames
vp2.render_anim(size=(600,400),
filename=fname,
range=(25,nframes),
renderer=renderer)try:
import google.colab
from IPython.display import Image
Image(open(fname, 'rb').read())
except ImportError:
"Assuming local run."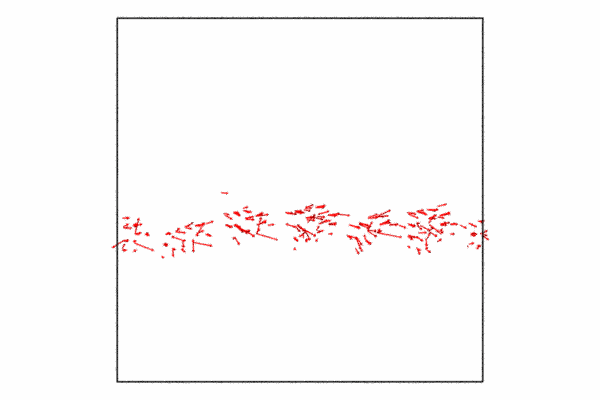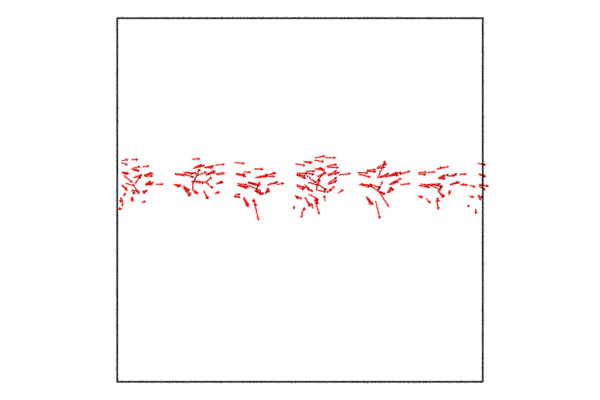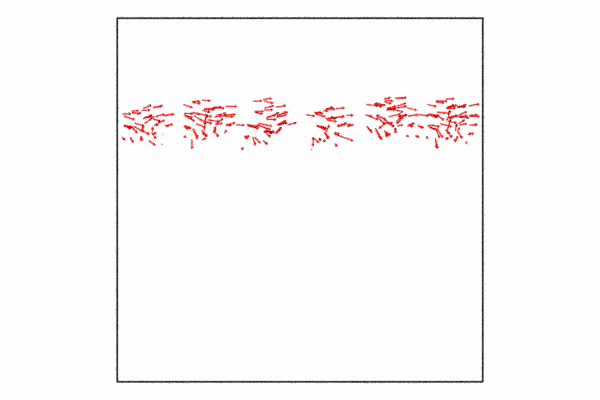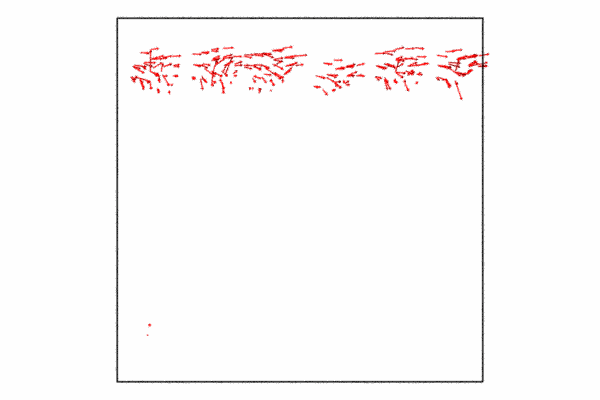
References
Citation
@online{bringuier2024,
author = {Bringuier, Stefan},
publisher = {Github Pages},
title = {Recipe 3: {Color} Coding Grain Boundary Atoms},
date = {2024-02-22},
url = {https://stefanbringuier.github.io/HowToSOVITO},
langid = {en}
}